Girvan-Newman algorithm
The Girvan-Newman algorithm for the detection and analysis of community structure relies on the iterative elimination of edges that have the highest number of shortest paths between nodes passing through them. By removing edges from the graph one-by-one, the network breaks down into smaller pieces, so-called communities. The algorithm was introduced by Michelle Girvan and Mark Newman.
How does it work?
The idea was to find which edges in a network occur most frequently between other pairs of nodes by finding edges betweenness centrality. The edges joining communities are then expected to have a high edge betweenness. The underlying community structure of the network will be much more fine-grained once the edges with the highest betweenness are eliminated which means that communities will be much easier to spot.
The Girvan-Newman algorithm can be divided into four main steps:
- For every edge in a graph, calculate the edge betweenness centrality.
- Remove the edge with the highest betweenness centrality.
- Calculate the betweenness centrality for every remaining edge.
- Repeat steps 2-4 until there are no more edges left.
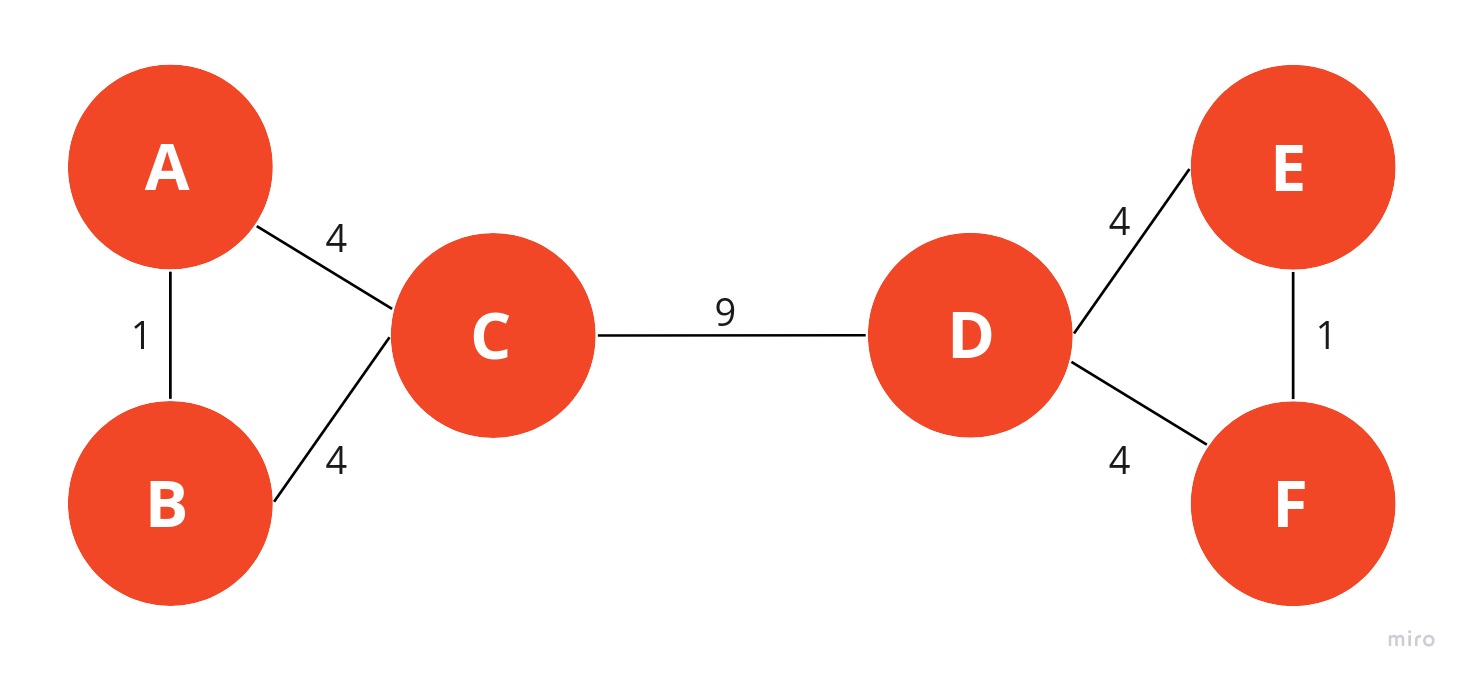
In this example, you can see how a typical graph looks like when edges are assigned weights based on the number of shortest paths passing through them. To keep things simple, we only calculated the number of undirected shortest paths that pass through an edge. The edge between nodes A and B has a strength of 1 because we don’t count A->B and B->A as two different paths.
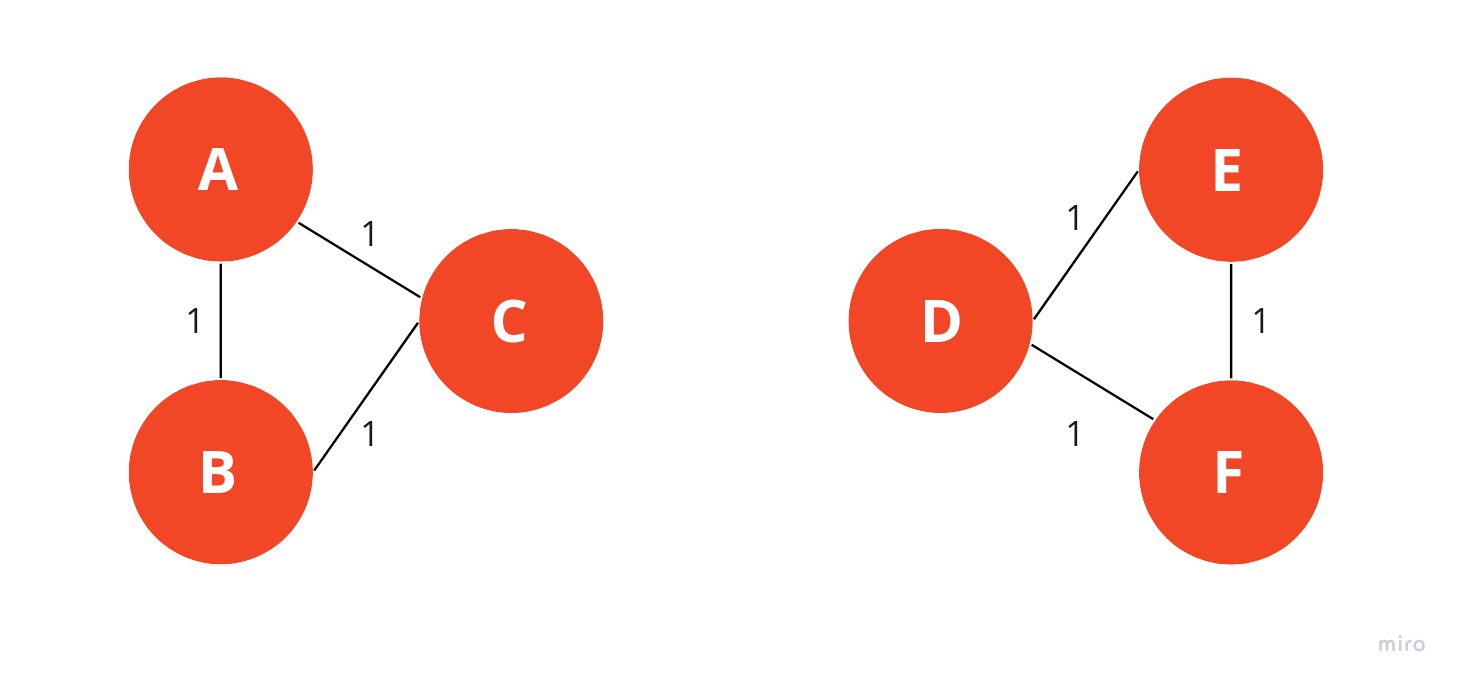
The Girvan-Newman algorithm would remove the edge between nodes C and D because it is the one with the highest strength. As you can see intuitively, this means that the edge is located between communities. After removing an edge, the betweenness centrality has to be recalculated for every remaining edge. In this example, we have come to the point where every edge has the same betweenness centrality.
Betweenness centrality
Betweenness centrality measures the extent to which a vertex or edge lies on paths between vertices. Vertices and edges with high betweenness may have considerable influence within a network by virtue of their control over information passing between others.
The calculation of betweenness centrality is not standardized and there are many ways to solve it. It is defined as the number of shortest paths in the graph that pass through the node or edge divided by the total number of shortest paths.
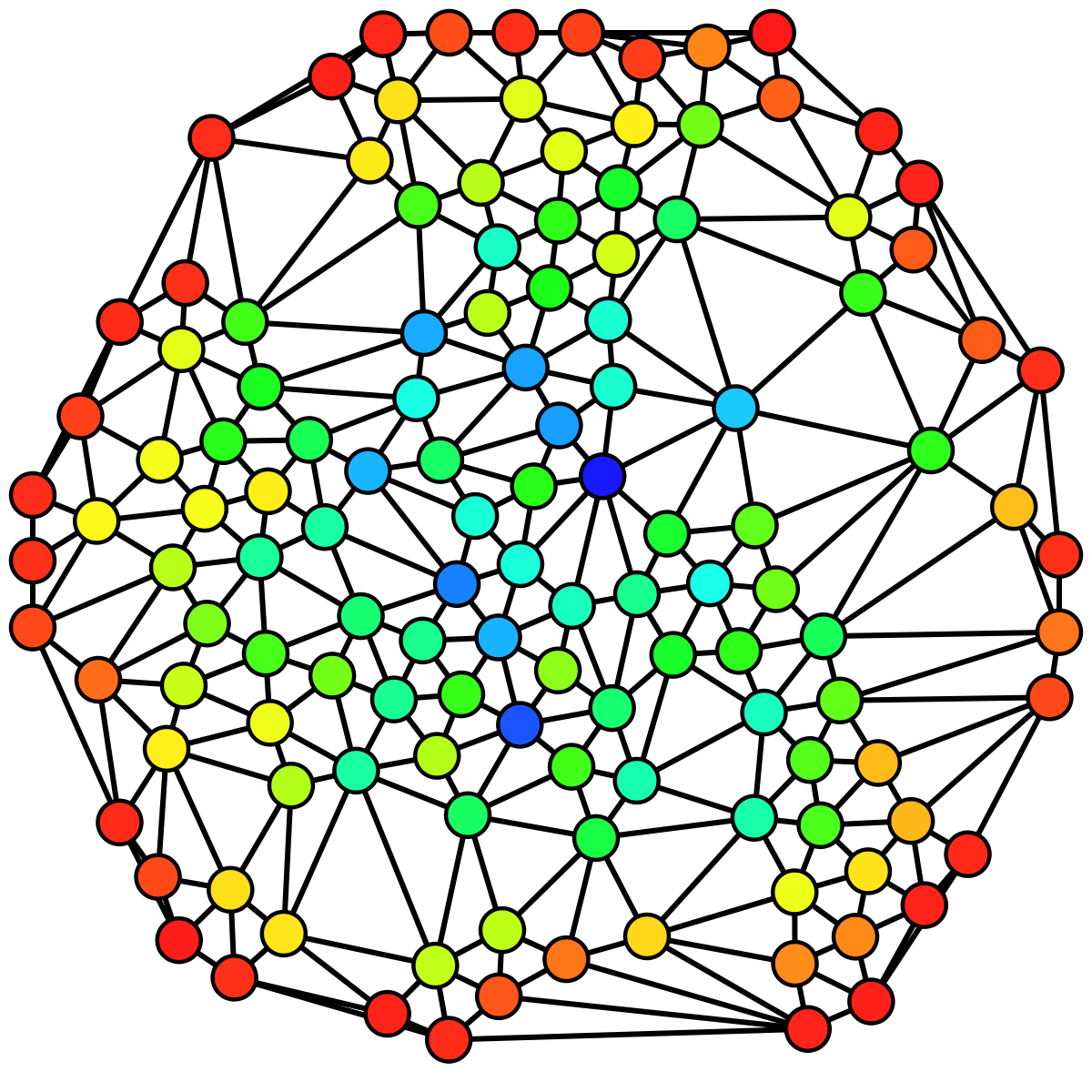
The image above shows an undirected graph colored based on the betweenness centrality of each vertex from least (red) to greatest (blue).
Read more about it at the betweenness centrality page.
Pseudocode
In each iteration, calculate the betweenness centrality for all edges in the graph. Remove the edge with the highest centrality. Repeat until there are no more edges left.
REPEAT
LET n BE number of edges in the graph
FOR i=0 to n-1
LET B[i] BE betweenness centrality of edge i
IF B[i] > max_B THEN
max_B = B[i]
max_B_edge = i
ENDIF
ENDFOR
remove edge i from graph
UNTIL number of edges in graph is 0
Usage in NetworkX
girvan_newman(G, most_valuable_edge=None)
Not fast enough? Find 100x faster algorithms here.
Method input
The first input parameter of the method, G, is a NetworkX graph.
The second parameter, most_valuable_edge, is a function that takes a graph as input and returns the edge that should be removed from the graph in each iteration. If no function is specified, the edge with the highest betweenness centrality will be chosen in each iteration.
Method output
The output of the method is an iterator over tuples of sets of nodes in G. Each set of nodes represents a community and each tuple is a sequence of communities at a particular level (iteration) of the algorithm.
Example
- Python code
- Output
- Visualization
import matplotlib.pyplot as plt
import networkx as nx
from networkx.algorithms.community.centrality import girvan_newman
G = nx.karate_club_graph()
communities = girvan_newman(G)
node_groups = []
for com in next(communities):
node_groups.append(list(com))
print(node_groups)
color_map = []
for node in G:
if node in node_groups[0]:
color_map.append("red")
else:
color_map.append("orange")
nx.draw(G, node_color=color_map, with_labels=True)
plt.show()
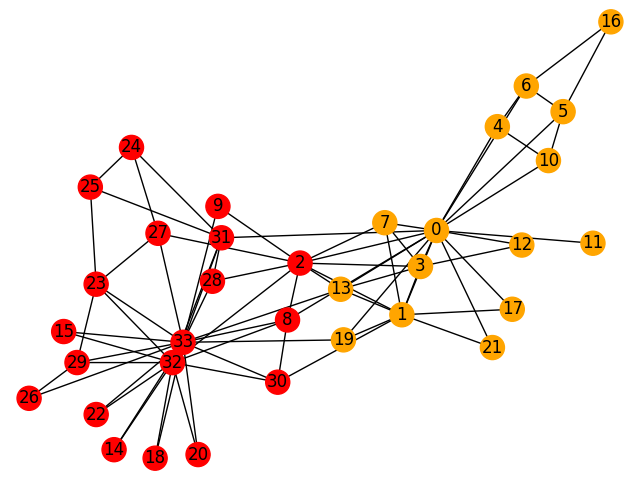
[[0, 1, 3, 4, 5, 6, 7, 10, 11, 12, 13, 16, 17, 19, 21],
[2, 8, 9, 14, 15, 18, 20, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33]]
The network has been divided into two distinct communities:
Where to next?
There are many graph algorithms libraries out there, with their own implementations of Girvan-Newman's algorithm. NetworkX's algorithms are written in Python, and there are many other libraries that offer faster C++ implementations, such as MAGE, a graph algorithms library developed by Memgraph team.