Weakly connected components algorithm (Union find)
A weakly connected component is a subgraph that is unreachable from other nodes/vertices of a graph or subgraph. The algorithm was described by A. Galler and Michael J. in 1964 and specific implementations either utilize breadth-first search or depth-first search to find the graph components. How does it work? The Weakly Connected Components algorithm (WCC), also known as Union Find, searches for distinct sets of connected nodes in a graph. All nodes in such a set are reachable from any other node in the same set. While the Strongly Connected Components algorithm (SCC) requires nodes to be reachable in both directions, WCC only requires nodes to be reachable in one direction. Both algorithms are used for network structure analysis.
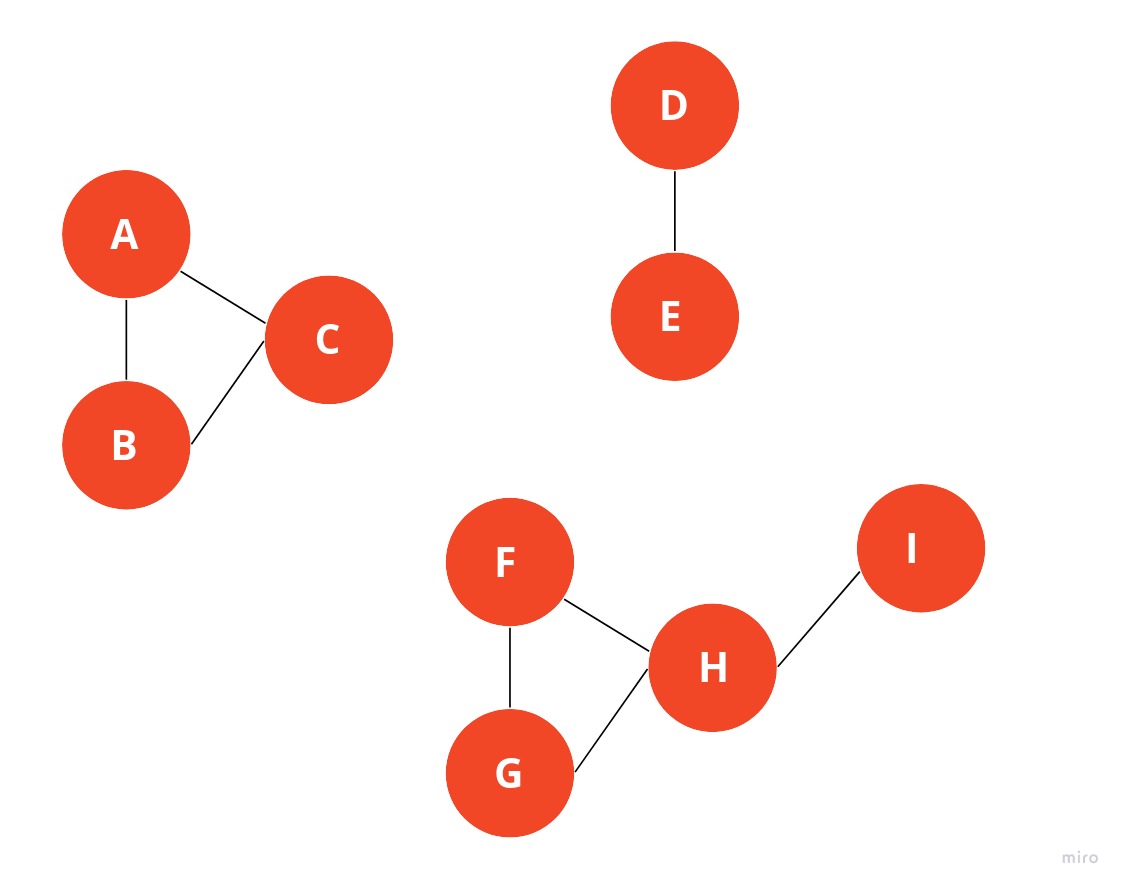
For example, in the graph above, you can see three weakly connected components. Nodes from the subgraph {A, B, C} aren’t connected to any other nodes in the graph and therefore must be a separate component.
Practical Applications
- The Weakly Connected Component algorithm is mostly used for graph pre-processing. Many graph algorithms require networks to be fully connected without distinct components. Union find can be used to find and potentially eliminate such components.
- WCC can be used for basic community detection use cases where distinct disconnected groups are expected.
- Some social networks can utilize weakly connected components for recommendation generating purposes.
Pseudocode
Start by labeling all nodes as unvisited. Then, iterate over the nodes in any order. For each node, if it wasn’t visited, run BFS from that node and add all reachable nodes as the same component. Add them to the list of visited nodes as well. If the node was already visited, skip it.
visited_nodes = []
components = []
FOR EACH node n in graph
IF n is not in visited_nodes THEN
connected_nodes = BFS(v)
ADD connected_nodes TO visited_nodes
ADD connected_nodes TO component
ENDIF
ENDFOR
Usage in NetworkX
weakly_connected_components(G)
Not fast enough? Find 100x faster algorithms here.
Method input
The input parameter of the method, G, is a directed graph.
Method output
The output of the method is a generator of sets. Each set contains the nodes of one weakly connected component.
Example
- Python code
- Output
- Visualization
import matplotlib.pyplot as plt
import networkx as nx
from networkx.algorithms.components import weakly_connected_components
G = nx.path_graph(4, create_using=nx.DiGraph())
nx.add_path(G, [10, 11, 12])
communities = weakly_connected_components(G)
node_groups = []
for com in communities:
node_groups.append(list(com))
print(node_groups)
color_map = []
for node in G:
if node in node_groups[0]:
color_map.append('orange')
else:
color_map.append('red')
nx.draw(G, node_color=color_map, with_labels=True)
plt.show()
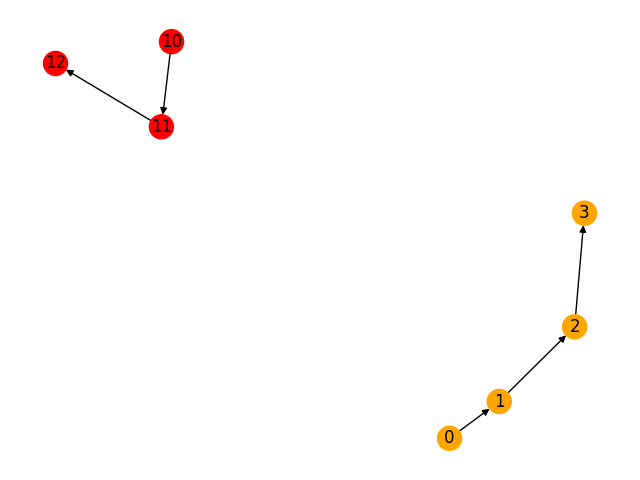
[[0, 1, 2, 3], [10, 11, 12]]
The network has two weakly connected components:
Where to next?
There are many graph algorithms libraries out there, with their own implementations of weakly connected components algorithm. NetworkX's algorithms are written in Python, and there are many other libraries that offer faster C++ implementations, such as MAGE, a graph algorithms library developed by Memgraph team.